Focus on Digital Medicine in Retina
Take-home points
|
 |
|
Bios Mr. Mishra is a research intern at Doheny Eye Institute, Pasadena, California, and graduate student at the Case Western Reserve University School of Medicine, Cleveland. Mr. Wang is a research intern at Doheny Eye Institute and an undergraduate student at the University of California Los Angeles School of Engineering. Dr. Saha is with the Doheny Eye Institute and has an additional appointment at the Commonwealth Scientific and Industrial Research Organisation (CSIRO) in Canberra, Australia. Dr. Sadda is director, artificial intelligence and imaging research, at Doheny Eye Institute and professor at UCLA David Geffen School of Medicine. Dr. Hu leads the Doheny Eye Image Analysis Laboratory at the Doheny Eye Institute. DISCLOSURES: Dr. Sadda disclosed relationships with Heidelberg Engineering, The other authors have no relevant relationships to disclose. Acknowledgment: Research reported here was partially supported by the National Eye Institute. |
An estimated 8 million people in the United States age 55 and older have some form of intermediate or advanced AMD1, 2 and automated techniques could play a critical role in the timely screening and detection of these patients. Ideally, AMD would be caught in its early stages before it progresses to irreversible vision loss due to complications such as outer retinal atrophy or exudative neovascular membranes.
It’s also critical to identify patients at high risk of progressing to advanced AMD, allowing for staging and establishing appropriate monitoring intervals. Early identification, accurate staging and risk stratification of early AMD stages could allow for prevention or possible early intervention, while also playing a role in facilitating the development of new treatments and preventative therapeutics.
Advanced AMD is defined by the hallmark features of central atrophy or macular neovascularization and commonly occurs with associated loss of vision. Patients with intermediate AMD have a 27 percent chance of progression to advanced AMD in five years, higher when the other eye is already afflicted with advanced AMD.3
Even with effective treatments for neovascular AMD, these patients still frequently lose vision due to the development of geographic atrophy, for no approved treatment yet exists, although some agents that may slow progression are under evaluation or regulatory assessment.4 This article explores the potential for automated imaging for the detection and monitoring of AMD.
Challenges with current methods
Historically, color fundus photography has been the gold standard for detecting and measuring GA. Fundus autofluorescence imaging is a noninvasive, in vivo two-dimensional imaging technique for metabolic mapping of naturally or pathologically occurring fluorophores of the ocular fundus.5 It has proved to be a useful method for identifying atrophic lesions due to its ability to provide high contrast.
In atrophy, photoreceptors and retinal pigment epithelium cells are lost, leading to the depletion of the fluorophores (in lipofuscin) and, consequently, reduced autofluorescence or hypofluorescence. Well-demarcated FAF hypofluorescence is the hallmark of atrophy. Studies of color fundus photos have identified several risk markers for AMD progression: large drusen; increased total drusen area; hyperpigmentation; and depigmentation.3,6,7
Recently, optical coherence tomography, due to its ability to provide three-dimensional cross-sectional anatomic information of retinal abnormalities, has gained favor over color fundus photography.8 Several novel OCT-based features have been identified as risk factors of AMD progression.9 They include higher central drusen volume,10 intraretinal hyperreflective foci,11 heterogeneous internal reflectivity within drusenoid lesions (IRDL),12 and reticular pseudodrusen (RPD) or subretinal drusenoid deposits (SDD).13,14,15 Unfortunately, while OCT provides many opportunities to further understand AMD biomarkers, it requires expert training and generates massive amounts of image data (up to hundreds of B-scans per examination). This limits the practicality of manual OCT analysis.
 |
|
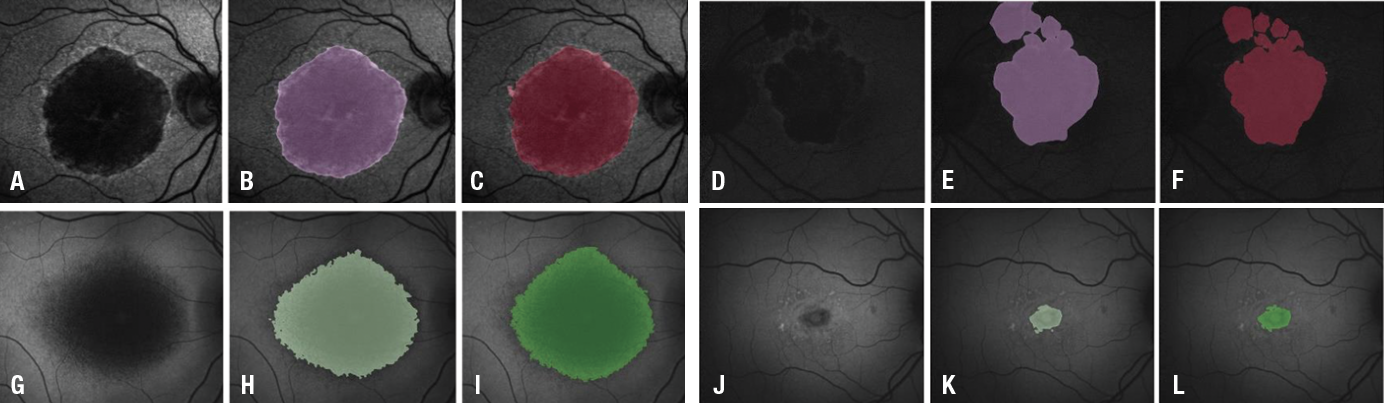
Figure 1: An illustration of segmentation system results. The upper row shows atrophic age-related macular degeneration segmentation: A and D fundus autofluorescence; B and E U-net segmentation; and C and F manual delineation. The lower row shows atrophic juvenile macular degeneration segmentation: G and J FAF images; H and K U-net segmentation; and I and L manual delineation. (Source: Proc. SPIE 10950, Medical Imaging 2019: Computer-Aided Diagnosis, 109501Q [13 March 2019; open source]). |
Automated image segmentation and analysis are desirable for both FAF and OCT. This can be accomplished while preserving performance and accuracy through deep learning techniques.
Late-stage screening and detection
Previously, our group at the Doheny Image Analysis Laboratory (DIAL) developed a fully convolutional neural network-based algorithm for atrophic AMD (or GA) screening and segmentation on FAF images.16 The U-net algorithm consists of the usual contracting network and expansive network,17 but supplements it through the use of upsampling operators and concatenating the high-resolution features of the contracting network to the upsampled features of the expansive network, leading to a more precise final output.
This algorithm has also been applied to the segmentation of Stargardt disease or atrophic juvenile macular degeneration (JMD). Additionally, researchers used deep residual convolutional neural networks to screen and distinguish atrophic eyes from normal eyes.18
The screening system algorithm our DIAL group developed demonstrated a high accuracy, with 0.98 for atrophic AMD and 0.95 for atrophic JMD against the manual gradings (Figure 1).16 The segmentation system algorithm also demonstrated a high overlapping ratio: 0.89 ± 0.06 for atrophic AMD and 0.78 ± 0.17 for atrophic JMD compared with the manual delineations. These highly robust systems indicate the great potential to facilitate large-scale atrophic AMD and JMD clinic trials, clinical research and translation to clinic daily application.
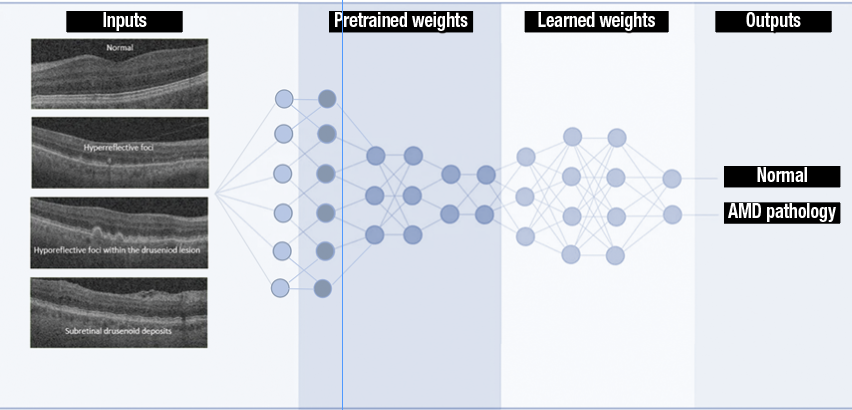 |
| Figure 2: Deep learning for identifying the presence of early age-related macular degeneration biomarkers employs transfer learning, with which state-of-the-art classification performance is achieved with relatively small datasets by using pretrained models to initiate network parameters, which are then fine-tuned by training on the desired image dataset. Neuron connections shown here are for illustration only. (Source: Sci Rep. 2019;9:10990 [open access]). |
Screening early AMD features
In 2019 our DIAL group reported on deep convolution neural networks trained to detect and classify several OCT B-scan features that had previously been found to be associated with AMD progression.19 They include hyperreflective foci, hyporeflective foci within the drusen and subretinal drusenoid deposits.9 The algorithm (Figure 2) employs transfer learning, with which state-of-the-art classification performance is achieved using relatively small datasets by using pretrained models to initiate network parameters, which are then fine-tuned by training on the desired image dataset.20 This allows for fast network training while avoiding over-fitting and ensuring robust performance. The algorithm also uses a U-net (i.e., ReLayNet21) to presegment a region of interest to increase performance of the model.
This deep-learning system achieved an overall accuracy of 87 percent for detecting and classifying the early AMD biomarkers identified. In the clinic, such a system could rapidly screen B-scans that require further investigation, improving the accuracy and efficiency of the diagnosis. The reported deep-learning system is one way to ease the burden of AMD on the health-care system, showing the potential to play a role in clinical decision support for patient management and screening.
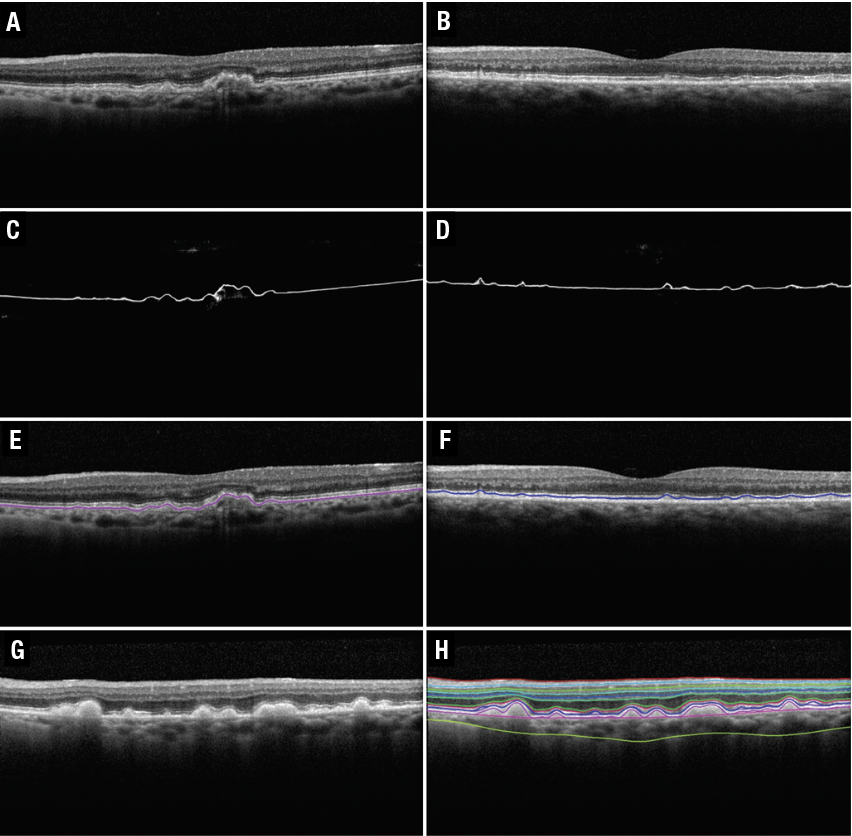 |
| Figure 3: Segmentation of regular drusen, reticular psuedodrusen (RPD), and 11 retinal layers on spectral-domain optical coherence tomography B-scans. A and B) Original B-scans for the segmentations of regular drusen and RPD shown in E and F, respectively. The drusen segmentation shown in E arises from the deep learning probability map shown in C. The RPD segmentation shown in F arises from the deep-learning probability map shown in D and G) Original B-scan for the 11 retinal-layer segmentation shown in H. (Source: Sci Rep. 2020;10:9541 [open access]) |
Detection of early AMD features
DIAL developed a deep-learning system to automatically segment drusen and RPD in SD-OCT images.22 The algorithm first uses a fully convolutional neural network based on the U-net to generate probability maps for the drusen and RPD present in each B-scan. Using these probability maps and the normalized gradient in the z-direction of the B-scan, a shortest-path algorithm automatically segmented both drusen and RPD. The neural network could also generate probability maps for the layers of the retina, making the what we described as the Deep Learning–Shortest Path algorithm generalizable to the segmentation of all retinal layers in addition to drusen and RPD.
The algorithm achieved a subpixel level mean difference for all retinal layers. For RPD in particular, mean and absolute mean differences between automated and manual segmentations were −0.75 ± 1.99 pixels (−2.92 ± 7.74 μm) and 1.53 ± 1.47 pixels (5.97 ± 5.74 μm), respectively. This study demonstrated that RPD, regular drusen and all retinal layers can be automatically and separately segmented on SD-OCT images (Figure 3). Quantifying and monitoring the progression of these features have significant potential for aiding in the further understanding of the evolution of AMD.
Bottom line
While no effective treatment for atrophic AMD currently exists, early identification before permanent vision loss occurs could allow staging and monitoring at appropriate intervals, which could open the door to new preventative treatments and early intervention studies. However, identifying early AMD biomarkers manually is time-consuming and requires expert training, often making it impractical or unfeasible.
Deep learning offers a potential solution to automatically screen for and detect biomarkers of both early and late AMD while preserving much of the accuracy of manual segmentation. The studies we reviewed are encouraging for the application of deep-learning methods in the screening and detection of AMD biomarkers for further clinical research, early intervention clinical trials and daily clinical use. RS
References
1. VanNewKirk MR, Nanjan MB, Wang JJ, Mitchell P, Taylor HR, McCarty CA. The prevalence of age related maculopathy: the visual impairment project. Ophthalmology. 2000;107:1593–1600 .
2. Bressler NM, Bressler SB, Congdon NG, et al. N.M. et al. Potential public health impact of age-related eye disease study results: AREDS report no. 11. Arch Ophthalmol. 2003;121:1621–1624.
3. Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436.
4. Jack LS, Sadiq MA, Do DV, Nguyen QD. Emixustat and lampalizumab: Potential therapeutic options for geographic atrophy. Dev Ophthalmol. 2016;55:302–309.
5. Schmitz-Valckenberg S, Holz FG, Bird AC, Spaide RF. Fundus autofluorescence imaging: review and perspectives, Retina. 2008;28:385–409.
6. Mitchell P, Smith W, Attebo K, Wang JJ. Prevalence of age-related maculopathy in Australia. The Blue Mountains Eye Study. Ophthalmology. 1995:102;1450–1460.
7. Klein R, Klein BE, Jensen SC, Meuer SM. The five-year incidence and progression of age-related maculopathy: The Beaver Dam Eye Study. Ophthalmology. 1997;104:7–21.
8. Drexler W, FujimotoJG. State-of-the-art retinal optical coherence tomography. Prog Retinal Eye Res. 2008;27:45–88.
9. Lei, J. et al. Proposal of a simple optical coherence tomography-based scoring system for progression of age-related macular degeneration. Graefe’s Archive Clin Experiment Ophthalmol. 2017;255:1551–1558.
10. Abdelfattah NS, Zhang H, Boyer DS, et al. Drusen volume as a predictor of disease progression in patients with late age-related macular degeneration in the fellow eye. Invest Ophthalmol Vis Sci. 2016;57:1839–1846 .
11. Nassisi M, Fan W, Shi Y, et al. Quantity of intraretinal hyperreflective foci in patients with intermediate age-related macular degeneration correlates with 1-year progression. Invest Ophthalmol Vis Sci. 2018:59:3431–3439.
12. Ouyang Y, Heussen FM, Hariri A, Keane PA, Sadda SR. Optical coherence tomography-based observation of the natural history of drusenoid lesion in eyes with dry age-related macular degeneration. Ophthalmology. 2013;120:2656–2665.
13. Finger RP, Wu Z, Luu CD, et al. Reticular pseudodrusen: a risk factor for geographic atrophy in fellow eyes of individuals with unilateral choroidal neovascularization. Ophthalmology. 2014;121: 1252–1256.
14. Marsiglia M, Boddu S, Bearelly S, et al. Association between geographic atrophy progression and reticular pseudodrusen in eyes with dry age-related macular degeneration. Invest Ophthalmol Vis Sci. 2013;54:7362–7369.
15. Zhou Q, Daniel E, Maguire MG, et al. Pseudodrusen and incidence of late age-related macular degeneration in fellow eyes in the comparison of age-related macular degeneration treatments trials. Ophthalmology. 2016;123: 1530–1540.
16. Wang Z, Sadda SR, Hu Z. Deep learning for automated screening and semantic segmentation of age-related and juvenile atrophic macular degeneration. Proc. SPIE 10950, Medical Imaging 2019: Computer-Aided Diagnosis, 109501Q (13 March 2019) https://doi.org/10.1117/12.2511538
17. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical Image Computing and Computer-Assisted Intervention– ICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham. 2015:234–241
18. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV: IEEE 770-778. doi:10.1109/CVPR.2016.90.
19. Saha S, Nassisi M, Wang M, et al. Automated detection and classification of early AMD biomarkers using deep learning. Sci Rep. 2019;9: 10990. https://doi.org/10.1038/s41598-019-47390-3
20. Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans Medical Imaging. 2016;35:1299-1312
21. Roy AG, Conjeti S, Karri SP, et al. ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed Optics Express. 2017;8:3627–3642.
22. Mishra Z, Ganegoda A, Selicha J, Wang Z, Sadda SR, Hu Z.Automated retinal layer segmentation using graph-based algorithm incorporating deep-learning-derived information. Sci Rep. 2020;10:9541.



